Genetic engineering is the direct manipulation of DNA to change an organism’s characteristics (phenotype) in a specific way. Scientists use genetic engineering to improve or modify an individual organism’s characteristics. Any organism, from a virus to a sheep, can be subjected to genetic engineering. Plants with higher nutritional value or resistance to herbicides, for example, can be produced using genetic engineering.
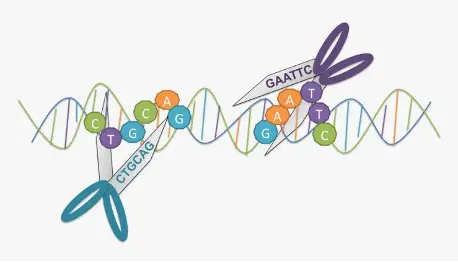
1. Restriction Enzymes (Molecular Scissor)
In protein engineering, the discovery of restriction endonucleases, and DNA-cleaving enzymes has been critical. Based on the nucleotide sequence, these enzymes cut DNA at specific locations. Hundreds of restriction enzymes capable of cutting DNA at a specific site have been isolated from various bacteria strains. When you cut DNA with a restriction enzyme, you get a lot of smaller fragments of varying sizes. Gel electrophoresis or chromatography can is used to separate these.
2. DNA Ligase
DNA ligase is a crucial enzyme in gene technology because it acts as a molecular suture, connecting two strands of DNA that require ATP. During joining, the enzyme forms a covalent bond between the 5′ phosphoryl group of one strand and the 3′ hydroxyl group of the neighboring strand. Two phosphodiester bonds are formed between two cohesive ends. The blunt ends produced by some enzymes inhibit the joining process. In such cases, joining necessitates a very high enzyme concentration. This enzyme is primarily obtain from the T4 phage virus.
3. Gel Electrophoresis
It will be pointless to purify DNA from cell culture or cut it with restriction enzymes if we couldn’t see the DNA – that is, if we couldn’t see if your extract contains anything or what size fragments you’ve cut it. However, there is a method to accomplish this- gel electrophoresis. Gels is used for various purposes, including detecting DNA inserts and knockouts and viewing cut DNA.

4. Methods to Select Transgenic Organisms f3-large
It is not required that all cells take up DNA during transformation. As a result, differentiating transformed cells from non-transformed cells is critical. Antibiotic resistance genes are commonly found in plasmids, and transgenic cells are selected based on the expression of those genes and their ability to grow in antibiotic-containing media. Other reporter proteins, such as the x-gal/lacZ system and the green fluorescence protein, allow for color and fluorescence selection.
5. Polymerases
Polymerases are groups of enzymes that catalyze the synthesis of nucleic acid molecules. The common name for the nucleic acid template on which the polymerase acts. The three major polymerases are listed below. DNA polymerase is a DNA-dependent DNA polymerase that replicates DNA from DNA. A reverse transcriptase (RNA-dependent DNA polymerase) that transcribes DNA from RNA. DNA-dependent RNA polymerase is a DNA-dependent RNA polymerase that transcribes RNA from DNA.
6. Reverse Transcriptase
It is an avian myeloblast virus-derived RNA-dependent DNA polymerase. The abbreviation for this is AMV-RT. A complementary DNA primer to the RNA template is what require by the enzyme. AMV-RT is made from two major parts. Reverse transcriptase is used in genetic engineering to create cDNA. Its job is to convert mRNA into single-stranded DNA.
7. Prokaryotic Host
The bacterium Escherichia coli was the first organism used in DNA technology experiments, and it is still many scientists’ preferred host. Without a doubt, E.coli, the most basic Gram-negative bacterium (a common bacterium of human and animal intestines), has played an important role in the evolution of modern biotechnology. E. coli can double every 20 minutes in the right conditions. As bacteria multiply, their plasmids (along with foreign DNA) multiply to produce millions of copies, known as a colony or clone. A clone is a collection of cells, organisms, or genes formed by multiplying a single cell, organism, or gene.

8. Eukaryotic Host
Because their complex structures (distinct organelles) are better suited to synthesising complex proteins, eukaryotic organisms are preferred to produce human proteins. Yeast is present. The most common eukaryotic organism is Saccharomyces cerevisiae. It is a non-pathogenic organism found in the brewing and baking industries. Gene cloning experiments use certain fungi.
9. Competent Host
Because DNA is a hydrophilic molecule, it cannot pass through cell membranes. Why? Before bacterial cells can be forced to take up the plasmid, they must first be considered “competent” to take up DNA. This is accomplished by giving the bacteria a specific amount of a divalent cation, such as calcium, which increases the efficiency of DNA entering the bacterium through openings or pores in the cell wall. Recombinant DNA can then be pushed into such cells by first incubating them on ice, exposing them to 42C for a time period (heat shock), and then returning them to ice. The bacteria can now digest the recombinant DNA. This is not the only method by which foreign DNA can enter host cells. Microinjection is a technique that involves injecting recombinant DNA directly into the nucleus of an animal cell.
10. Selection of Small Self-Replicating DNA
Plasmids are small circular pieces of DNA that do not belong to a bacterial genome but can self-replicate. Plasmids are frequently used as gene transfer vectors between microorganisms. Once the desired gene has been amplified and the gene and the plasmid have been cut by restriction enzymes, they are ligated together to form recombinant DNA. Vectors can be viral (bacteriophage) DNA or cosmids, recombinant plasmids containing bacteriophage genes.