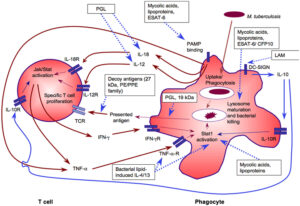
Despite being the challenge of extensive research, tuberculosis, resulting from Mycobacterium tuberculosis, stays at the gift the main motive of loss of life from an infectious agent. Secreted and molecular wall proteins have interaction with the host and play vital roles in pathogenicity. These proteins are explored as candidate diagnostic markers, ability drug goals or vaccine antigens, and extra currently unique interest is being given to the position in their post-translational adjustments.
1. Mycobacterial Pressure And Boom Conditions
Mycobacterium tuberculosis H37Rv pressure (ATCC® 25618™) changed into grown for three weeks at 37°C in Lowenstein Jensen stable medium and after boom changed into accomplished it changed into subculture in Middlebrook 7H9 broth supplemented with albumin, dextrose, and catalase (ADC) enrichment (Difco, Detroit, MI, USA) for 12 days with mild agitation at 37°C. Mycobacterial cells had been pelleted at 4000xg for 15 min at 4°C and washed three instances with bloodless phosphate-buffered saline. Mycobacterial cells had been finally cultured as floor pellicles for three to four weeks at 37°C without shaking in 250 mL of Sauton minimum medium, an artificial protein-unfastened tradition medium, which changed into organized as formerly defined.
2. Culture Filtrate Protein Preparation
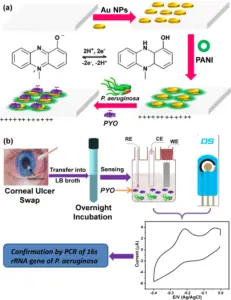
Bacterial cells had been eliminated with the aid of using centrifugation and traditional filtrate protein (CFP) changed into organized with the aid of using filtering the supernatant via 0.2 μM pore length filters (Millipore, USA). After sterility trying out of CFP in Mycobacteria Growth Indicator Tube (MGIT) supplemented with MGIT 960 supplement (BD, Bactec) for forty-two days at 37°C in BD BACTEC™ MGIT™ computerized mycobacterial detection system, CFP changed into focused the usage of centrifugal clear out devices (Macrosep Advance, 3kDa MWCO (Pall Corporation, USA)).
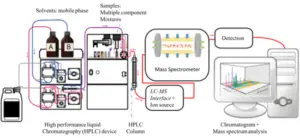
3. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
Two replicas of M. tuberculosis CFP (25 μg) had been loaded in SDS-PAGE 15% and stained with CBB G-250 as defined elsewhere [23]. Six gel slices had been excised from every lane in line with protein density. In-gel Cys alkylation, in gel-digestion and peptide extraction, changed into done as defined before [24]. Tryptic peptides had been separated the usage of nano-HPLC (Ultimate 3000, Thermo Scientific) coupled online with a Q-Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fischer Scientific).
4. Protein Evaluation
Identified proteins in every mirror had been compared with the aid of using area-proportional Venn Diagram evaluation (BioVenn [29]) and a listing of not unusual place proteins changed into generated. The further evaluation took into consideration proteins found in each replicate of LC-MS/MS evaluation. SEPro module retrieved a listing of proteins recognized with Uniprot code.
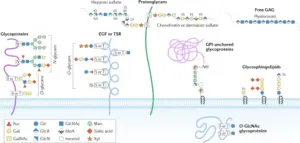
5. Protein O-Glycosylation Evaluation
Proteins bearing O-glycosylated peptides in each replicates had been as compared with the aid of using area-proportional Venn Diagram evaluation and a listing of not unusual place glycosylated proteins for every of the analyzed adjustments, i.e. Hex, Hex-Hex, and Hex-Hex-Hex, changed into generated. Further evaluation changed into manually done to perceive not unusual place changed peptides within side the listing of not unusual place glycosylated proteins, in addition to not unusual place adjustments. As a result of this evaluation, a listing of proteins with no unusual place adjustments changed into generated, consisting of proteins having the identical changed peptide in each replicates.
6. Estimation Of Protein Abundance And Comparative Evaluation
To estimate protein abundance Normalized Spectral Abundance Factor (NSAF) was calculated with Pattern Lab for proteomics software program changed into taken into consideration. NSAF permits for the estimation of protein abundance with the aid of using dividing the sum of spectral counts for every recognized protein with the aid of using its length, for this reason figuring out the spectral abundance factor (SAF), and normalizing this fee in opposition to the sum of the overall protein SAFs within side the pattern. Proteins had been ordered in line with their NSAF.
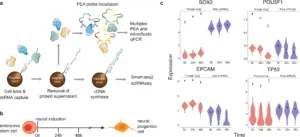
7. Protein Category
Gene Ontology evaluation of the traditional filtrate proteins changed into done with David Gene Functional Classification Tool the usage of the Cellular Component Ontology database and overall proteins of M. tuberculosis H37Rv as background. With this evaluation, predominant classes of enriched phrases for P75%, P90%, P95%, and overall proteins had been determined. Functional category of traditional filtrate proteins changed into done in line with useful classes of M. tuberculosis database Myco browser.
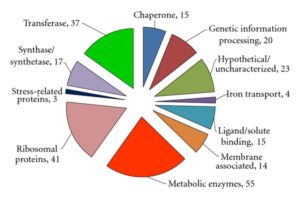
8. O-Glycosylation Validation
The identical analytical workflow defined for O-glycosylation evaluation of our records changed into done the usage of the uncooked records documents deposited on the ProteomeXchange Consortium with the dataset identifier PXD000111. This evaluation changed into done to examine the changed peptides recognized in our paintings in opposition to extra organic replicates acquired in preceding paintings that substantially characterized traditional filtrate proteins of M. tuberculosis H37Rv.
9. Protein Category Is The Usage Of A Quality-Quantitative Evaluation
Quantitative proteomics primarily based totally on spectral counting techniques are trustworthy to appoint and were proven to effectively hit upon variations among samples. To do not forget the pattern-to-pattern version acquired whilst wearing out mirror analyses, and the truth that longer proteins tend to have extra peptide identifications than shorter proteins, Pattern lab for Proteomics software program makes use of NSAF (Normalized spectral abundance factor) for spectral counting normalization.
10. Conclusion
Membrane and exported proteins are important gamers for the protection and survival of bacterial organisms in inflamed hosts, and their contribution to pathogenesis and immunological responses make those proteins applicable goals for biomedical research. Consistently, diverse of the proteins recognized in M.